
Get the full story of the microbiome
- Importance of Microbial Diversity
- Dysbiosis and Recurrent C. difficile Infection
- Historic Microbiome Restoration Approaches
- Patient and Healthcare Burdens
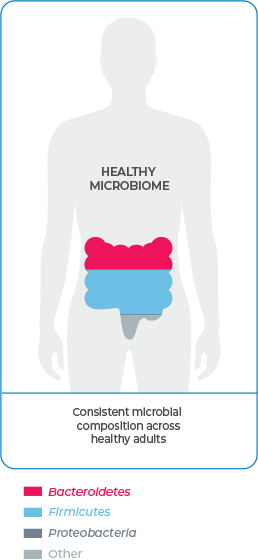
A diverse microbial composition is essential to overall health
The gut microbiome—the microbial community in the intestinal tract—is an influencer of metabolism and immunity and a mediator of resistance to some pathogenic infections.1,2
A distinct and essential organ within the body, the gut microbiome contains an estimated 500-1000 species and 100 trillion organisms, encoding 100-fold more unique genes than our own genome.1-4
The microbiome influences metabolism and immunity and acts as a mediator of resistance to some pathogenic infections.1
In its balanced state, there is a symbiotic relationship between luminal bacteria and our human cells.5
These cells communicate and form long-lasting, interactive associations that play a vital role in the conservation of mucosal immune function, epithelial barrier integrity, motility, and nutrient absorption.2,5,6
Bacteroidetes and Firmicutes are the most prevalent phyla in the gut microbiome and—working symbiotically—are vital to maintaining overall health and inhibiting C. difficile infection (CDI).1-3,7-9
Bacteroidetes (Gram-negative)
Their ability to adapt and persist in changing gut environments allows abundance and stability, providing long-term associations with human hosts and enabling functions that include7,10-12:
- Immunomodulatory effects7
- Inhibitory activities against C. difficile13
- Reduction of C. difficile colonization13
- Essential contributor to the stability of the microbiome7
Firmicutes (Gram-positive)
Composed of helpful and harmful bacteria, Firmicutes are the most abundant and diverse bacterial gut species with functions that include3,14:
- Anti-inflammatory effects15-17
- Fortification of gut barrier (along with other bacteria) observed preclinically18-20
- Most abundant phyla in the gut microbiome3
Proteobacteria
- A biomarker for inflammatory disease as well as C. difficile infection and colonization. CDI is associated with its increase21-24
Deficiencies in Bacteroidetes and Firmicutes are specifically associated with dysbiosis.3,5
THE BURDEN OF CDI MAY INCREASE
WHEN THE INFECTION RECURS
Can the power of the microbiome be
unlocked to break the cycle of rCDI?
Share this with colleagues!

References
- Gilbert JA, Blaser MJ, Caporaso JG, et al. Current understanding of the human microbiome. Nat Med. 2018;24(4):392-400.
- Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474(11):1823-1836.
- Antharam VC, Li EC, Ishmael A, et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol. 2013;51(9):2884-2892.
- Marchesi JR, Adams DH, Fava F, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65(2):330-339.
- Bien J, Palagani V, Bozko P. The intestinal microbiota dysbiosis and Clostridium difficile infection: is there a relationship with inflammatory bowel disease? Therap Adv Gastroenterol. 2013;6(1):53-68.
- Ley R, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647-1651.
- Wexler AG, Goodman AL. An insider’s perspective: Bacteroides as a window into the microbiome. Nat Microbiol. 2017;2(1):17026.
- Rinninella E, Raoul P, Cintoni M, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14.
- Nishijima S, Suda W, Oshima K, et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016;23(2):125-133.
- Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355-1359.
- Kurokawa K, Itoh T, Kuwahara T, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14(4):169-181.
- Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439.
- Li X, Kang Y, Huang Y, et al. A strain of Bacteroides thetaiotaomicron attenuates colonization of Clostridioides difficile and affects intestinal microbiota and bile acids profile in a mouse model. Biomed Pharmacother. 2021;137:111290.
- Kho ZY, Lal SK. The human gut microbiome—a potential controller of wellness and disease. Front Microbiol. 2018;9:1835.
- Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731-16736.
- Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139(6):1844-1854.e1.
- Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63(8):1275-1283.
- Paust S, Lu L, Mccarty N, Cantor H. Engagement of B7 on effector T cells by regulatory T cells prevents autoimmune disease. Proc Natl Acad Sci U S A. 2004;101(28):10398-10403.
- El Aidy S, van Baarlen P, Derrien M, et al. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5(5):567-579.
- Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138(1):1-11.
- Gu W, Li W, Wang W, et al. Response of the gut microbiota during the Clostridioides difficile infection in tree shrews mimics those in humans. BMC Microbiol. 2020;20(1):260-273.
- Qiu J, Zhou H, Jing Y, Dong C. Association between blood microbiome and type 2 diabetes mellitus: a nested case-control study. J Clin Lab Anal. 2019;33(4):e22842.
- Vestor-Anderson MK, Mirsepasi-Lauridsen HC, Prosberg MV, et al. Increased abundance of proteobacteria in aggressive Crohn’s disease seven years after diagnosis. Sci Rep. 2019;9(1):13473.
- Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131(20):346-352.e1-3.
